Online first
Current issue
Archive
Special Issues
About the Journal
Publication Ethics
Anti-Plagiarism system
Instructions for Authors
Instructions for Reviewers
Editorial Board
Editorial Office
Contact
Reviewers
All Reviewers
2024
2023
2022
2021
2020
2019
2018
2017
2016
General Data Protection Regulation (RODO)
RESEARCH PAPER
Disruption of mitochondrial function augments the radiosensitivity of prostate cancer cell lines
1
Medical University, Lublin, Poland
2
Independent Medical Biology Unit, Lublin, Poland
3
Department of Biochemistry and Molecular Biology, Medical University, Lublin, Poland
4
Human Anatomy Department, Medical University, Lublin, Poland
5
Department of Toxicology, Medical University, Lublin, Poland
Corresponding author
Ann Agric Environ Med. 2023;30(1):65-76
KEYWORDS
TOPICS
ABSTRACT
Introduction:
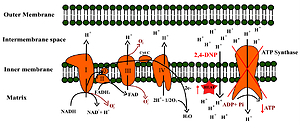
Ionizing radiation is one of the most widely used therapeutic methods in the treatment of prostate cancer, but the problem is developing radioresistance of the tumour. There is evidence that metabolic reprogramming in cancer is one of the major contributors to radioresistance and mitochondria play a crucial role in this process.
Objective:
The aim of the study was to assess the influence of oxidative phosphorylation uncoupling to radiosensitivity of prostate cancer cells differing in metabolic phenotype.
Material and methods:
LNCaP, PC-3 and DU-145 cells were exposed to X-rays and simultaneously treated with 2,4-dinitrophenol (2,4-DNP). The radiosensitive of cell lines was determined by cell clonogenic assay and cell cycle analysis. The cytotoxic effect was evaluated with MTT and CVS (Crystal violet staining) assay, apoptosis detection and cell cycle analysis. The phenotype of the cells was determined by glucose uptake and lactate release, ATP level measurement as well as basal reactive oxygen species level and mRNA expression of genes related to oxidative stress defence.
Results:
The synergistic effect of 2,4-dinitrophenol and X-ray was observed only in the case of the LNCaP cell line.
Conclusions:
Phenotypic analysis indicates that this may be due to the highest dependence of these cells on oxidative phosphorylation and sensitivity to disruption of their redox status.
Ionizing radiation is one of the most widely used therapeutic methods in the treatment of prostate cancer, but the problem is developing radioresistance of the tumour. There is evidence that metabolic reprogramming in cancer is one of the major contributors to radioresistance and mitochondria play a crucial role in this process.
Objective:
The aim of the study was to assess the influence of oxidative phosphorylation uncoupling to radiosensitivity of prostate cancer cells differing in metabolic phenotype.
Material and methods:
LNCaP, PC-3 and DU-145 cells were exposed to X-rays and simultaneously treated with 2,4-dinitrophenol (2,4-DNP). The radiosensitive of cell lines was determined by cell clonogenic assay and cell cycle analysis. The cytotoxic effect was evaluated with MTT and CVS (Crystal violet staining) assay, apoptosis detection and cell cycle analysis. The phenotype of the cells was determined by glucose uptake and lactate release, ATP level measurement as well as basal reactive oxygen species level and mRNA expression of genes related to oxidative stress defence.
Results:
The synergistic effect of 2,4-dinitrophenol and X-ray was observed only in the case of the LNCaP cell line.
Conclusions:
Phenotypic analysis indicates that this may be due to the highest dependence of these cells on oxidative phosphorylation and sensitivity to disruption of their redox status.
ACKNOWLEDGEMENTS
Internal Grant for young researchers of Medical University
of Lublin (PBmb10)
REFERENCES (49)
1.
Weir HK, Anderson RN, Coleman King SM, et al. Heart Disease and Cancer Deaths – Trends and Projections in the United States, 1969–2020. Prev Chronic Dis. 2016;13:E157. doi:10.5888/pcd13.160211.
2.
Cancer (IARC) TIA for R on. Global Cancer Observatory. Accessed April 15, 2022. https://gco.iarc.fr/.
3.
Parker C, Gillessen S, Heidenreich A, Horwich A, ESMO Guidelines Committee. Cancer of the prostate: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26 Suppl 5:v69–77. doi:10.1093/annonc/mdv222.
4.
Bolus NE. Basic Review of Radiation Biology and Terminology. J Nucl Med Technol. 2017;45(4):259–264. doi:10.2967/jnmt.117.195230.
5.
Burgio E, Piscitelli P, Migliore L. Ionizing Radiation and Human Health: Reviewing Models of Exposure and Mechanisms of Cellular Damage. An Epigenetic Perspective. Int J Environ Res Public Health. 2018;15(9):E1971. doi:10.3390/ijerph15091971.
6.
Fabbrizi MR, Parsons JL. Radiotherapy and the cellular DNA damage response: current and future perspectives on head and neck cancer treatment. Cancer Drug Resist. 2020;3(4):775–790. doi:10.20517/cdr.2020.49.
7.
Huang RX, Zhou PK. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Sig Transduct Target Ther. 2020;5(1):60. doi:10.1038/s41392-020-0150-x.
8.
McCann E, O’Sullivan J, Marcone S. Targeting cancer-cell mitochondria and metabolism to improve radiotherapy response. Transl Oncol. 2021;14(1):100905. doi:10.1016/j.tranon.2020.100905.
9.
Tang L, Wei F, Wu Y, et al. Role of metabolism in cancer cell radio-resistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37(1):87. doi:10.1186/s13046-018-0758-7.
10.
Potter M, Newport E, Morten KJ. The Warburg effect: 80 years on. Biochem Soc Trans. 2016;44(5):1499–1505. doi:10.1042/BST20160094.
11.
Liu C, Jin Y, Fan Z. The Mechanism of Warburg Effect-Induced Chemoresistance in Cancer. Front Oncol. 2021;11:698023. doi:10.3389/fonc.2021.698023.
12.
DeBerardinis RJ, Chandel NS. We need to talk about the Warburg effect. Nat Metab. 2020;2(2):127–129. doi:10.1038/s42255-020-0172-2.
13.
Vasan K, Werner M, Chandel NS. Mitochondrial Metabolism as a Target for Cancer Therapy. Cell Metab. 2020;32(3):341–352. doi:10.1016/j.cmet.2020.06.019.
14.
Lynam-Lennon N, Maher SG, Maguire A, et al. Altered Mitochondrial Function and Energy Metabolism Is Associated with a Radioresistant Phenotype in Oesophageal Adenocarcinoma. PLOS ONE. 2014;9(6): e100738. doi:10.1371/journal.pone.0100738.
15.
Priolo C, Pyne S, Rose J, et al. AKT1 and MYC induce distinctive metabolic fingerprints in human prostate cancer. Cancer Res. 2014;74(24):7198–7204. doi:10.1158/0008-5472.can-14-1490.
16.
Yang L, Hou Y, Yuan J, et al. Twist promotes reprogramming of glucose metabolism in breast cancer cells through PI3K/AKT and p53 signaling pathways. Oncotarget. 2015;6(28):25755–25769. doi:10.18632/oncotarget.4697.
17.
Geisler JG. 2,4 Dinitrophenol as Medicine. Cells. 2019;8(3):E280. doi:10.3390/cells8030280.
18.
Rui L. New Antidiabetes Agent Targeting Both Mitochondrial Uncoupling and Pyruvate Catabolism: Two Birds With One Stone. Diabetes. 2019;68(12):2195–2196. doi:10.2337/dbi19-0024.
20.
Geraldo de Campos E, Fogarty M, Spinosa De Martinis B, Kerr Logan B. Analysis of 2,4-Dinitrophenol in Postmortem Blood and Urine by Gas Chromatography-Mass Spectrometry: Method Development and Validation and Report of Three Fatalities in the United States. J Forensic Sci. 2020;65(1):183–188. doi:10.1111/1556-4029.14154.
21.
Kuznetsov AV, Javadov S, Sickinger S, Frotschnig S, Grimm M. H9c2 and HL-1 cells demonstrate distinct features of energy metabolism, mitochondrial function and sensitivity to hypoxia-reoxygenation. Biochim Biophys Acta. 2015;1853(2):276–284. doi:10.1016/j.bbamcr.2014.11.015.
22.
Zhao Q, Sun Q, Zhou L, Liu K, Jiao K. Complex Regulation of Mitochondrial Function During Cardiac Development. JAHA. 2019;8(13):e012731. doi:10.1161/JAHA.119.012731.
23.
Wheaton WW, Weinberg SE, Hamanaka RB, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;3:e02242. doi:10.7554/eLife.02242.
24.
Sena P, Mancini S, Benincasa M, Mariani F, Palumbo C, Roncucci L. Metformin Induces Apoptosis and Alters Cellular Responses to Oxidative Stress in Ht29 Colon Cancer Cells: Preliminary Findings. Int J Mol Sci. 2018;19(5):E1478. doi:10.3390/ijms19051478.
25.
Shrestha R, Johnson E, Byrne FL. Exploring the therapeutic potential of mitochondrial uncouplers in cancer. Mol Metab. 2021;51:101222. doi:10.1016/j.molmet.2021.101222.
26.
Demine, Renard, Arnould. Mitochondrial Uncoupling: A Key Controller of Biological Processes in Physiology and Diseases. Cells. 2019;8(8):795. doi:10.3390/cells8080795.
27.
Jiang H, Jin J, Duan Y, et al. Mitochondrial Uncoupling Coordinated With PDH Activation Safely Ameliorates Hyperglycemia via Promoting Glucose Oxidation. Diabetes. 2019;68(12):2197–2209. doi:10.2337/db19-0589.
28.
Vier J, Gerhard M, Wagner H, Häcker G. Enhancement of death-receptor induced caspase-8-activation in the death-inducing signalling complex by uncoupling of oxidative phosphorylation. Mol Immunol. 2004;40(10):661–670. doi:10.1016/j.molimm.2003.09.008.
29.
Han YH, Kim SW, Kim SH, Kim SZ, Park WH. 2,4-dinitrophenol induces G1 phase arrest and apoptosis in human pulmonary adenocarcinoma Calu-6 cells. Toxicol In Vitro. 2008;22(3):659–670. doi:10.1016/j.tiv.2007.12.005.
30.
Sun Y, St Clair DK, Fang F, et al. The radiosensitization effect of parthenolide in prostate cancer cells is mediated by nuclear factor-kappaB inhibition and enhanced by the presence of PTEN. Mol Cancer Ther. 2007;6(9):2477–2486. doi:10.1158/1535-7163.MCT-07-0186.
31.
van Bokhoven A, Varella-Garcia M, Korch C, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57(3):205–225. doi:10.1002/pros.10290.
32.
Josson S, Xu Y, Fang F, Dhar SK, St Clair DK, St Clair WH. RelB regulates manganese superoxide dismutase gene and resistance to ionizing radiation of prostate cancer cells. Oncogene. 2006;25(10):1554–1559. doi:10.1038/sj.onc.1209186.
33.
Scott SL, Gumerlock PH, Beckett L, Li Y, Goldberg Z. Survival and cell cycle kinetics of human prostate cancer cell lines after single- and multifraction exposures to ionizing radiation. Int J Radiat Oncol Biol Phys. 2004;59(1):219–227. doi:10.1016/j.ijrobp.2004.01.027.
34.
Zdrowowicz M, Datta M, Rychłowski M, Rak J. Radiosensitization of PC3 Prostate Cancer Cells by 5-Thiocyanato-2 '-deoxyuridine. Cancers. 2022;14(8):2035. doi:10.3390/cancers14082035.
35.
Humpton TJ, Vousden KH. Regulation of Cellular Metabolism and Hypoxia by p53. Cold Spring Harb Perspect Med. 2016;6(7):a026146. doi:10.1101/cshperspect.a026146.
36.
Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20(4):199–210. doi:10.1038/s41580–019–0110-x.
37.
Simabuco FM, Morale MG, Pavan ICB, Morelli AP, Silva FR, Tamura RE. p53 and metabolism: from mechanism to therapeutics. Oncotarget. 2018;9(34):23780–23823. doi:10.18632/oncotarget.25267.
38.
Higgins LH, Withers HG, Garbens A, et al. Hypoxia and the metabolic phenotype of prostate cancer cells. Biochim Biophys Acta. 2009;1787(12):1433–1443. doi:10.1016/j.bbabio.2009.06.003.
39.
Vaz CV, Alves MG, Marques R, et al. Androgen-responsive and nonresponsive prostate cancer cells present a distinct glycolytic metabolism profile. Int J Biochem Cell Biol. 2012;44(11):2077–2084. doi:10.1016/j.biocel.2012.08.013.
40.
Pertega-Gomes N, Felisbino S, Massie CE, et al. A glycolytic phenotype is associated with prostate cancer progression and aggressiveness: a role for monocarboxylate transporters as metabolic targets for therapy. J Pathol. 2015;236(4):517–530. doi:10.1002/path.4547.
41.
Effert P, Beniers AJ, Tamimi Y, Handt S, Jakse G. Expression of glucose transporter 1 (Glut-1) in cell lines and clinical specimens from human prostate adenocarcinoma. Anticancer Res. 2004;24(5A):3057–3063.
42.
Cutruzzola F, Giardina G, Marani M, et al. Glucose Metabolism in the Progression of Prostate Cancer. Front Physiol. 2017;8:97. doi:10.3389/fphys.2017.00097.
43.
Lim HW, Hong S, Jin W, et al. Up-regulation of defense enzymes is responsible for low reactive oxygen species in malignant prostate cancer cells. Exp Mol Med. 2005;37(5):497–506. doi:10.1038/emm.2005.62.
44.
Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2(1):17. doi:10.1186/2049–3002–2–17.
45.
Martinez-Outschoorn UE, Trimmer C, Lin Z, et al. Autophagy in cancer associated fibroblasts promotes tumor cell survival: Role of hypoxia, HIF1 induction and NF?B activation in the tumor stromal microenvironment. Cell Cycle. 2010;9(17):3515–3533. doi:10.4161/cc.9.17.12928.
46.
Sznarkowska A, Kostecka A, Meller K, Bielawski KP. Inhibition of cancer antioxidant defense by natural compounds. Oncotarget. 2017;8(9):15996–16016. doi:10.18632/oncotarget.13723.
47.
Bing Z, Yang G, Zhang Y, et al. Proteomic analysis of effects by x-rays and heavy ion in HeLa cells. Radiology and Oncology. 2014;48(2):142–154. doi:10.2478/raon-2013–0087.
48.
van Gisbergen MW, Zwilling E, Dubois LJ. Metabolic Rewiring in Radiation Oncology Toward Improving the Therapeutic Ratio. Front Oncol. 2021;11:653621. doi:10.3389/fonc.2021.653621.
49.
Nile DL, Rae C, Walker DJ, et al. Inhibition of glycolysis and mitochondrial respiration promotes radiosensitisation of neuroblastoma and glioma cells. Cancer Metab. 2021;9(1):24. doi:10.1186/s40170–021–00258–5.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.